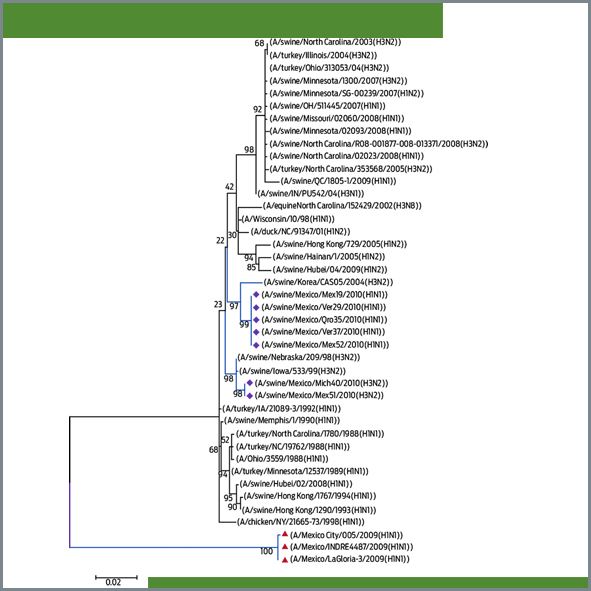
Genetic changes detected in the internal genes of porcine influenza viruses isolated in Mexico
Main Article Content
Abstract
Article Details
References
Ali A.; Khatri M, Wang L, Saif YM, and Lee CW. (2012). Identification of swine H1N2/pandemic H1N1 reassortant influenza virus in pigs, United States. Vet Microbiol.158, 60-8.
CDC. Update: drug susceptibility of swine-origin influenza A (H1N1) viruses, April 2009. Morbidity and Mortality Weekly. Report 58, 433-435.
Chen, GW. and Shih SR, (2009). Genomic signatures of influenza A pandemic (H1N1) 2009 virus. Emerg. Infect. Dis. 15, 1897–1903.
Dawood, F.S.; Jain, S., Finelli, L., Shaw, M.W., Lindstrom, S., Garten, R.J., Gubareva, L.V., Xu, X., Bridges, C.B. and Uyeki, T.M. (2009). Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 360, 2605–2615.
Escalera, Z.M.; Cobian-Guemes, G, Soto del Rio, M, Isa, P, Sanchez, B.I, Parissi, C.A, Martinez, C.M, Romero, P, Velazquez, S.L, Huerta, L.B, Nelson, M, Montero H, Vinuesa, P, Lopez, S, and Arias, C. (2012). Characterization of an influenza A virus in Mexican swine that is related to the A/H1N1/2009 pandemic clade. Virology 433, 176–182.
Gao, R. et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N. Engl. J. Med. 368, 1888–1897 (2013).
Garten, R.J.; Davis, C.T., Russell, C.A., Shu B, Lindstrom S, Balish A, Sessions WM, Xu X, Skepner E, Deyde V, Okomo-Adhiambo M, Gubareva L, Barnes J, Smith CB, Emery SL, Hillman MJ, Rivailler P, Smagala J, de Graaf M, Burke DF, Fouchier RA, Pappas C, Alpuche-Aranda CM, López-Gatell H, Olivera H, López I, Myers CA, Faix D, Blair PJ, Yu C, Keene KM, Dotson PD Jr, Boxrud D, Sambol AR, Abid SH, St George K, Bannerman T, Moore AL, Stringer DJ, Blevins P, Demmler-Harrison GJ, Ginsberg M, Kriner P, Waterman S, Smole S, Guevara HF, Belongia EA, Clark PA, Beatrice ST, Donis R, Katz J, Finelli L, Bridges CB, Shaw M, Jernigan DB, Uyeki TM, Smith DJ, Klimov A.I. and Cox NJ. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. (2009). Science. 325(5937):197-201
Gramer, M.R. (2008). An update on swine influenza ecology and diagnostics. Proceeding, 39th Annual Meeting of the American Association of Swine Veterinarians. March 8-11, 2008, San Diego, CA, USA.
Karasin, A.I.; Landgraf, J., Swenson, S., Erickson, G., Goyal, S., Woodruff, M.,Scherba, G., Anderson, G. and Olsen, C.W. (2002). Genetic characterization of H1N2 influenza A viruses isolated from pigs throughout the UnitedStates. J. Clin. Microbiol. 40, 1073–1079.
Karasin, A.I.; Carman, S and Olsen CW. (2006). Identification of human H1N2 and human-swine reassortant H1N2 and H1N1 influenza A viruses among pigs in Ontario, Canada (2003 to 2005). J Clin Microbiol, 44, 1123-1126.
Katz, J.M., Lu, X., et al., 2000. Molecular correlates of influenza A H5N1 virus pathogenesis in mice. Journal of Virology 74 (22), 10807–10810.
Kazutaka K, Kazuharu M, Kei-ichi K, Takashi M. (2002). “MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform”. Nucleic Acids Research 30 (14): 3059–66. doi:10.1093/nar/gkf436. PMC 135756. PMID 12136088.
Li, H, Ruan, J, Durbin, R. (2008). Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18, 1851–1858.
Mehle A.; Dugan, V, Taubenberger, J, and Doudna, J. (2011). Reassortment and Mutation of the Avian Influenza Virus Polymerase PA Subunit Overcome Species Barriers Journal of Virology. 1750–1757.
Moreno, A; Trani, D.L, Alborali, L, Vaccari, G, Barbieri, I, Falcone, E, Sozzi, E, Puzelli, S, Ferri, G, and Cordioli P. (2010). First pandemic H1N1 outbreak from a pig farm in Italy. Open Virol. J. 4, 52–56
Pan C.; Cheung B, Tan S, Li C, Li L, Liu S, and Jiang S, (2009). Genomic signature and mutation trend analysis of pandemic (H1N1) 2009 influenza A virus. PLoS One 5, e9549.
Pereda, A.; Rimondi, A, Cappuccio, J, Sanguinetti, R, Angel, M, Ye J, Sutton, T, Diba ́rbora M, Olivera, V, Craig, MI, Quiroga, M, Machuca, M, Ferrero, A, Perfumo, C, and Perez DR. (2011). Evidence of reassortment of pandemic H1N1 influenza virus in swine in Argentina: are we facing the expansion of potential epicenters of influenza emergence? Influenza Other Respir. Viruses 5, 409–412.
Posada D. (2008). JModeltest: Phylogenetic Model Averaging. Molecular Biology and Evolution 25: 1253-1256.
Rott R.; Orlich M, and Scholtissek C. (1979). Correlation of pathogenicity and gene constellation of influenza A viruses. Non-pathogenic recombinants derived from highly pathogenic parent strains. J. Gen. Virol. 44, 471-477.
Scholtissek C.; Stech J, Krauss S, and Webster RG, (2002). Cooperation between the hemagglutinin of avian viruses and the matrix protein of human influenza A viruses. J. Virol. 76, 1781–1786.
Scholtissek, C.; Buerger, H., Kistner, O, and Shortridge K. F. (1985). The nucleoprotein as a possible major factor in determining hosts specificity of influenza H3N2 viruses. Virology 147, 287-294.
Seo S, H, Hoffmann E, Webster R, G (2002) Lethal H5N1 influenza viruses escape host anti-viral cytokine responses. Nat Med 8: 950–954.
Shu, B.; Garten, R, Emery, S, Balish A, Cooper, L, Sessions, W, Deyde, V, Smith C, Berman, L, Klimov, A, Lindstrom, S, and Xu X. (2012). Genetic analysis and antigenic characterization of swine origin influenza viruses isolated from humans in the United States, 1990–2010. Virology 422. 151–160.
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics, 22, 2688–2690.
Tamura, K, Peterson, D, Peterson, N, Stecher, G, Nei M, and Kumar, S. (2011). MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance and Maximum Parsimony Methods. Mol. Biol. Evol. 28, 2731–2739.
Tang J.W, Shetty N, Lam T.T, Hon K.L. (2010). Emerging, novel, and known influenza virus infections in humans. Infect Dis Clin North Am. 24:603-617.
Team, R. (2010). A language and Environment for Statistical Computing, R Foundation for Statistical Computing Vienna.
Tong S, Zhu X, Li Y, Shi M, Zhang J, et al. (2013) New World Bats Harbor Diverse Influenza A Viruses. PLoS Pathog 9(10): e1003657. doi:10.1371/ journal.ppat.1003657.
Vincent, A.L.; Ma, K.M. Lager, B.H. Janke, and J.A. Richt. (2008). Swine Influenza Viruses a North American perspective. Adv. Virus. Res. 72,127-54.
Vincent A.L,; Swenson, S.L., Lager K.M, Gauger, P.C, Loiacono, C, and Zhang Y. (2009). Characterization of an influenza A virus isolated from pigs during anoutbreak of respiratory disease in swine and people during acounty fair in the United States. Veterinary Microbiology 137, 51–59.
Watanabe T, Kawaoka Y. (2011) Pathogenesis of the 1918 Pandemic Influenza Virus. PLoS Pathog 7(1): e1001218. doi:10.1371/journal.ppat.1001218.
Webster R.; Bean W, Gorman O, Chambers T, and Kawaoka Y. (1992). Evolutionand Ecology of Influenza A Viruses. Microbiological Reviews. 56, 152-179.
Webby, R.J.; Rossow, K., Erickson, G., Sims, Y. and Webster, R., (2004). Multiplelineages of antigenically and genetically diverse influenza Avirus co-circulate in the United States swine population. Virus. Res.103, 67–73.
Weingartl, H.M.; Berhane, Y, Hisanaga, T, Neufeld, J, Kehler, H, Emburry-Hyatt C, Hooper-McGreevy, K, Kasloff, S, Dalman, B, Bystrom, J, Alexandersen, S, Li Y, and Pasick J. (2010). Genetic and pathobiologic characterization of pandemic H1N1 2009 influenza viruses from a naturally infected swine herd. J Virol. 84: 2245-56.
Zhou, N.N.; Senne, D.A, and Landgraf, J.S. (1999). Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J Virol, 73, 8851-8856.
License

Veterinaria México OA by Facultad de Medicina Veterinaria y Zootecnia - Universidad Nacional Autónoma de México is licensed under a Creative Commons Attribution 4.0 International Licence.
Based on a work at http://www.revistas.unam.mx
- All articles in Veterinaria México OA re published under the Creative Commons Attribution 4.0 Unported (CC-BY 4.0). With this license, authors retain copyright but allow any user to share, copy, distribute, transmit, adapt and make commercial use of the work, without needing to provide additional permission as long as appropriate attribution is made to the original author or source.
- By using this license, all Veterinaria México OAarticles meet or exceed all funder and institutional requirements for being considered Open Access.
- Authors cannot use copyrighted material within their article unless that material has also been made available under a similarly liberal license.